install.packages("INLA",
repos=c(getOption("repos"),
INLA="https://inla.r-inla-download.org/R/testing"), dep=TRUE)Data Simulation
This section describes the steps to simulate occurrence data for fitting spatially explicit occupancy models in R-INLA.
Make sure you have that latest R and R-INLA versions installed:
Rversion4.4.0or above-
R-INLAversionINLA_24.06.04or above. For latest testing version use:
1 Set up
Define the spatial domain and the regular lattice by setting a \(300 \times 300\) study area divided into \(3\times3\) grid cells.
Code
# Define spatial domain
win <- owin(c(0,300), c(0,300))
npix <- 1000
Domain <- rast(nrows=npix, ncols=npix,
xmax=win$xrange[2],xmin=win$xrange[1],
ymax = win$yrange[2],ymin=win$yrange[1])
values(Domain) <- 1:ncell(Domain)
xy <- crds(Domain)
# Define regular grid
cell_size = 3
customGrid <- st_make_grid(Domain,cellsize = c(cell_size,cell_size)) %>%
st_cast("MULTIPOLYGON") %>%
st_sf() %>%
mutate(cellid = row_number())
# number of cells
ncells <- nrow(customGrid)
2 Simulate data for a simple spatial occupancy model
We simulate three different Gaussian Markov Random fields (GMRF) using the inla.qsample function.
Code
# Spatial boundary
boundary_sf = st_bbox(c(xmin = 0, xmax = 300, ymax = 0, ymin = 300)) %>%
st_as_sfc()
# Create a fine mesh
mesh_sim = fm_mesh_2d(loc.domain = st_coordinates(boundary_sf)[,1:2],
offset = c(-0.1, -.2),
max.edge = c(4, 50))
# Matern model
matern_sim <- inla.spde2.pcmatern(mesh_sim,
prior.range = c(100, 0.5),
prior.sigma = c(1, 0.5))
range_spde = 100
sigma_spde = 1
# Precision matrix
Q = inla.spde.precision(matern_sim, theta = c(log(range_spde),
log(sigma_spde)))
# Simulate three spatial fields
seed = 12345
sim_field = inla.qsample(n = 3, Q = Q, seed = seed)Create the projector matrix A_proj using the centroid of each grid cell as coordinates and compute the spatial random effect \(\omega(s)\), spatial environmental covariate \(x(s)\) and spatial detection covariate \(g_1(s)\) (Figure 2). (Note: Covariates \(x(s)\) and \(g_1(s)\) are assumed to be known and are stored as raster data).
Code
# Obtain the centroid of each cell
coord_grid = st_coordinates(customGrid %>% st_centroid())
# A matrix
A_proj = inla.spde.make.A(mesh_sim, loc = coord_grid)
# Spatial components
omega_s = (A_proj %*% sim_field)[,1] # spatial random field
x_s = (A_proj %*% sim_field)[,2] # spatial environmental covariate
g_s = (A_proj %*% sim_field)[,3] # spatial detection covariate
# create rasters
x_rast = rast(data.frame(x = coord_grid[,1], y = coord_grid[,2],x_s))
g_rast = rast(data.frame(x = coord_grid[,1], y = coord_grid[,2],g_s))
# save raster data
writeRaster(x_rast,filename = paste('raster data','x_covariat.tif',sep='/'),
overwrite=TRUE)
writeRaster(g_rast,filename = paste('raster data','g_covariat.tif',sep='/'),overwrite=TRUE)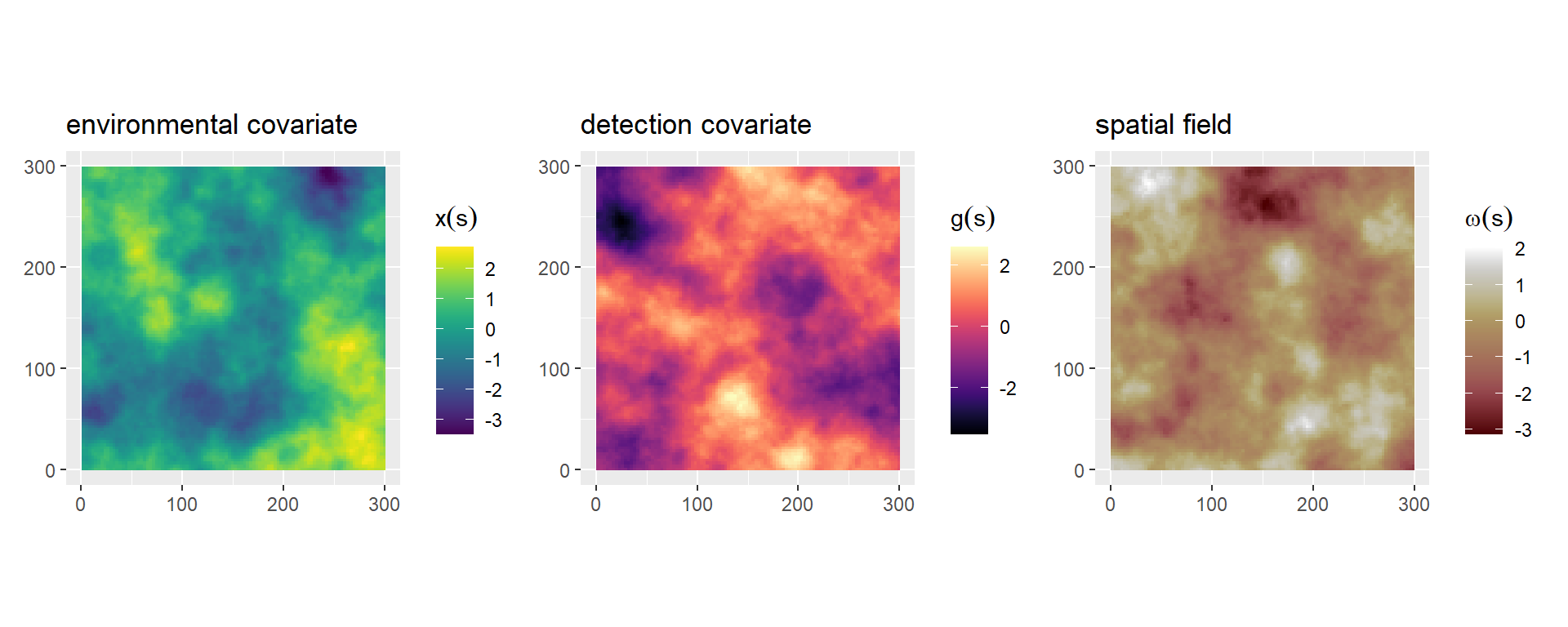
Occurrence data is simulated from the following occupancy model:
\[\begin{align} \mbox{State process:}& \begin{cases} z_i \sim \mathrm{Bernoulli}(\psi_i) \nonumber \\\mathrm{logit}(\psi_i) = \beta_0 + \beta_1 \ x_i(s) + \omega_i(s) \\ \end{cases}\\ \mbox{ Observational process:}& \begin{cases} y_{ij} \sim \mathrm{Bernoulli}(z_i \times p_{ij}) \nonumber \\\mathrm{logit}(p_{ij}) = \alpha_0 + \alpha_1 \ g_{1i}(s) + \alpha_2 \ g_{2ij}\\ \end{cases} \end{align}\]Here, \(g_{2}\) is a survey-level covariate that allows detection probabilities to vary between visits/surveys ( see Section 2.2 for simulation details).
2.1 State process model sub-component
Define model parameters \(\beta\) and compute the occupancy probabilities \(\psi\) and occupancy states \(z\) (Figure 3)
Code
# State process model coeficcients
beta <- c(NA,NA)
beta[1] <- qlogis(0.3) # Base line occupancy probability
beta[2] <- 1.5 # environmental covariate effect
# Occupancy probabilities
psi <- inla.link.logit(beta[1] + beta[2]*x_s + omega_s, inverse = T)
# True occupancy state
set.seed(seed)
z <- rbinom(ncells, size = 1, prob = psi)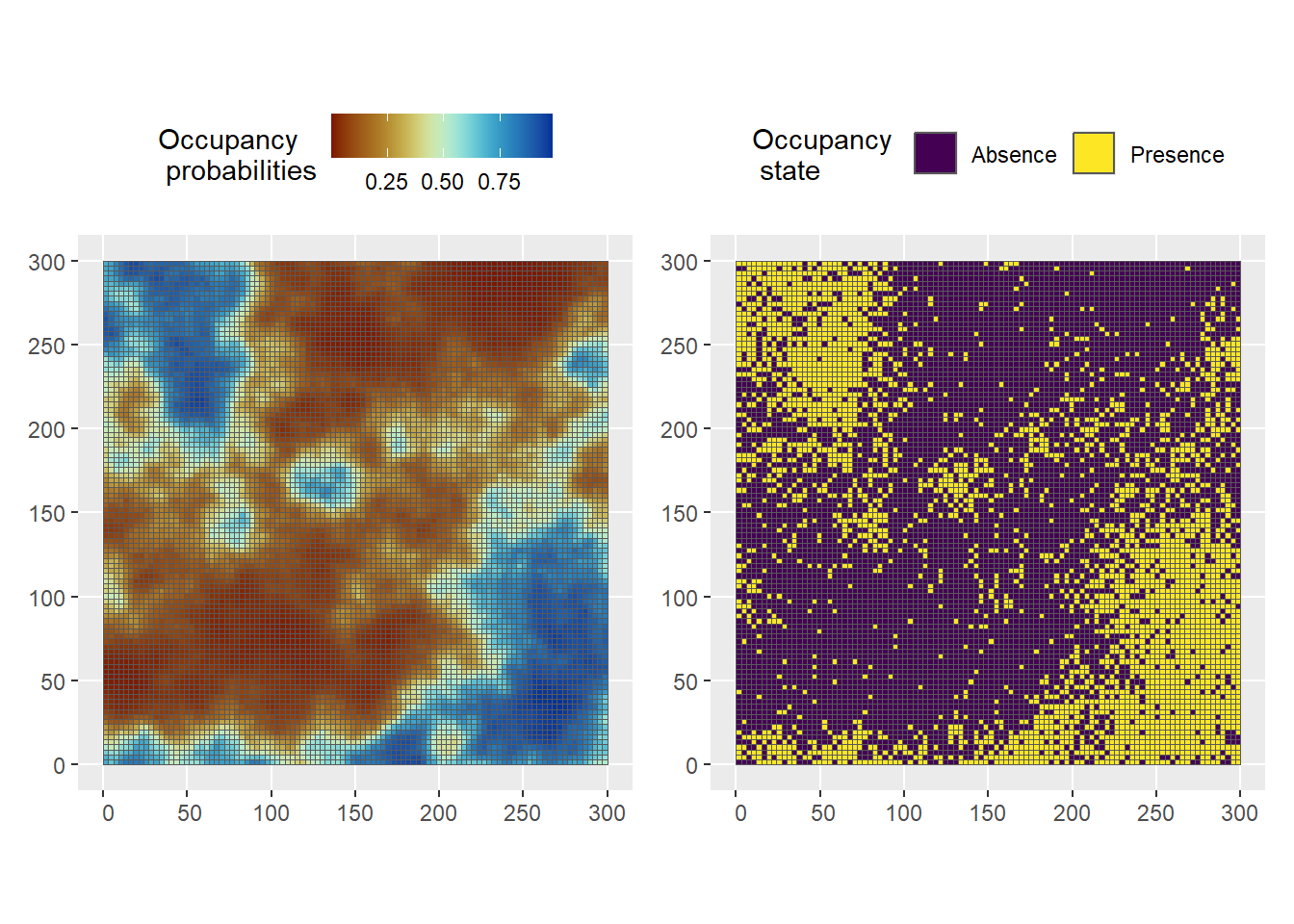
2.2 Observational process model sub-component
Firs, a random sample of 20 % of the total cells is obtained.
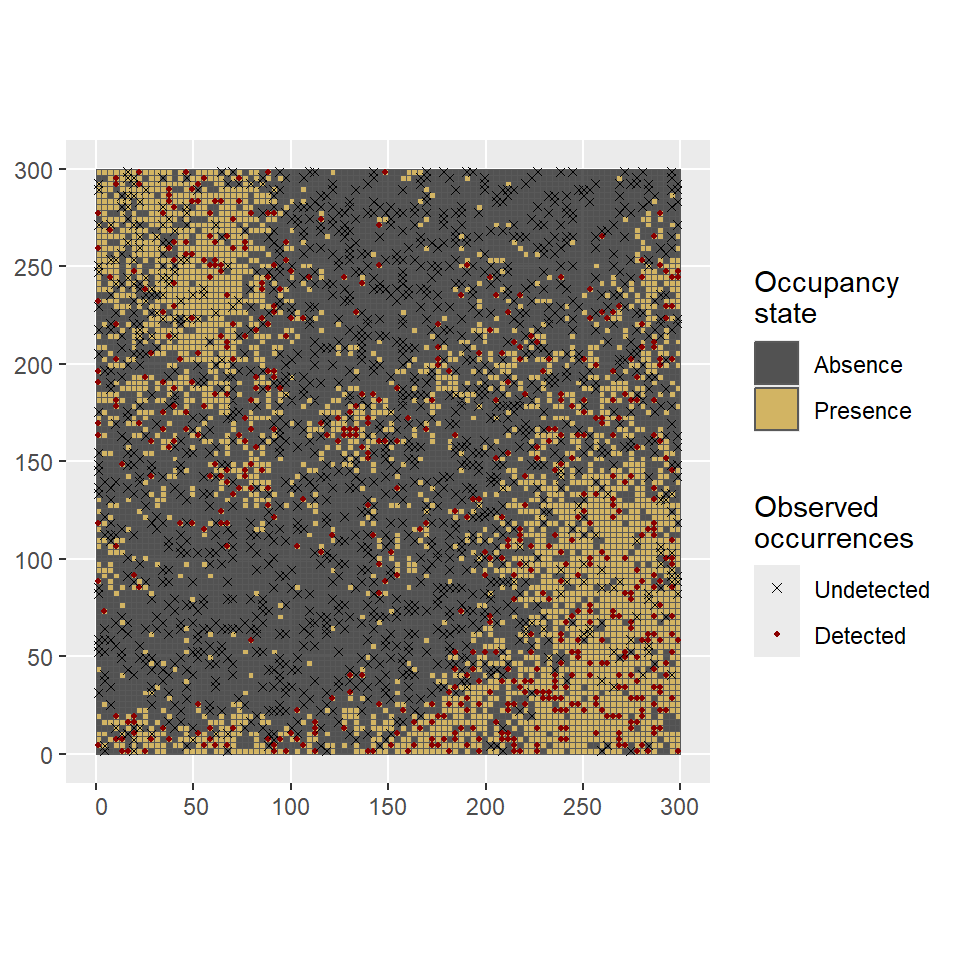
Then, we define model parameters \(\alpha\) , detection probabilities \(p\) and observed number of occurrences \(y\) across \(K=3\) visits per site/cell (Figure 4). Additionally, a survey-level covariate \(g_2\) is simulated at every surveyed cell to allow detection probabilities to vary by visit.
Code
# Number of vistis
K= 3
# Observational process model coeficcients
alpha <- c(NA,NA,NA)
alpha[1] <- qlogis(0.6) # Base line detection probability
alpha[2] <- 1 # detection covariate g1 effect
alpha[3] <- 0.5 # detection covariate g2 effect
# Survey-level detection covariate g2
g2 <- array(runif(n = nsites * K, -1, 1), dim = c(nsites, K))
# Detection probabilities and observed occurrences
# Create empty matrix to store the results
y <- p.mat <- matrix(NA,nrow = nsites,ncol=3)
# loop over visits
for(j in 1:K){
p.mat[,j] <- inla.link.logit(alpha[1] +
alpha[2]*g_s[site_id] +
alpha[3]*g2[, j], inverse = T)
y[,j] <- rbinom(n = nsites,size = 1,prob = p.mat[,j]*z[site_id] )
}Create data set Table 1:
Code
# centroid of the cell
Occ_data_1 <- customGrid %>%
st_centroid() %>%
filter(sample==1) %>%
dplyr::select(-c('sample'))
Obs_data <- data.frame(y = y, # detectio/non-detection data
g2 = g2, # survey level covariate
cellid = site_id)
Occ_data_1 <- left_join(Occ_data_1,Obs_data,by = "cellid")
# append coordinates as columns
Occ_data_1[,c('x.loc','y.loc')] <- st_coordinates(Occ_data_1)
Occ_data_1 = st_drop_geometry(Occ_data_1)
# Save CSV file for analysis
write.csv(Occ_data_1,file='Occ_data_1.csv',row.names = F)| cellid | y.1 | y.2 | y.3 | g2.1 | g2.2 | g2.3 | x.loc | y.loc |
|---|---|---|---|---|---|---|---|---|
| 2 | 0 | 0 | 0 | 0.5714531 | 0.3866185 | -0.2529522 | 4.5 | 1.5 |
| 5 | 0 | 0 | 1 | -0.1941702 | -0.7073133 | 0.4481461 | 13.5 | 1.5 |
| 6 | 0 | 0 | 1 | -0.3740240 | 0.9385314 | -0.1100978 | 16.5 | 1.5 |
| 7 | 0 | 0 | 0 | 0.3056664 | 0.5610409 | 0.7647462 | 19.5 | 1.5 |
| 9 | 1 | 0 | 0 | 0.6081176 | -0.4150512 | 0.1087413 | 25.5 | 1.5 |
| 23 | 0 | 0 | 0 | 0.1278117 | -0.9585974 | -0.9708703 | 67.5 | 1.5 |
3 Simulate space-time occupancy data
Here we simulate data for a space-time occupancy model by defining a SPDE for the spatial domain and an autoregressive model of order 1, i.e. AR(1), for the time component (this is separable space-time model where the spatio-temporal covariance is defined by the Kronecker product of the covariances of the spatial and temporal random effects, see (Cameletti et al. 2012) for more details).
We set a total of nT = 5 discrete time points where the occupancy state changes. We can use the book.rspde() function available in the spde-book-functions.R file (Krainski et al. 2018) to simulate \(t = 1,\ldots,nT\) independent realizations of the random field1. Then, temporal correlation is introduced:
\[ \omega(s_i,t) = \rho \ \omega(s_i,t-1) + \epsilon(s_i,t), \]
where \(\pmb{\epsilon} \sim N(0,\Sigma = \sigma_{\epsilon}^2\tilde{\Sigma})\) are the spatially correlated random effects with Matérn correlation function \(\tilde{\Sigma}\) and \(\rho=0.7\) is the autoregressive parameter (\(\sqrt{1-\rho^2}\) is included so \(\omega(s_i,1)\) comes from a \(N(0,\Sigma/(1-\rho^2)\) stationary distribution; see (Cameletti et al. 2012)).
Code
# load helping functions
source('spde-book-functions.R')
# Time points
nT = 5
# parameters of the Matern
# Create a fine mesh
mesh_sim = fm_mesh_2d(loc.domain = st_coordinates(boundary_sf)[,1:2],
offset = c(-0.1, -.2),
max.edge = c(4, 50))
# Matern model
params <- c(sigma = sigma_spde, range = range_spde)
# generate samples from the random field
epsilon.t <- book.rspde(coord_grid, range = params[2], seed = seed,
sigma = params[1], n = nT, mesh = mesh_sim,
return.attributes = TRUE)
# temporal dependency
rho <- 0.65
omega_st <- epsilon.t
for (t in 2:nT){
omega_st[, t] <- rho * omega_st[, t - 1] + sqrt(1 - rho^2) * epsilon.t[, t]
}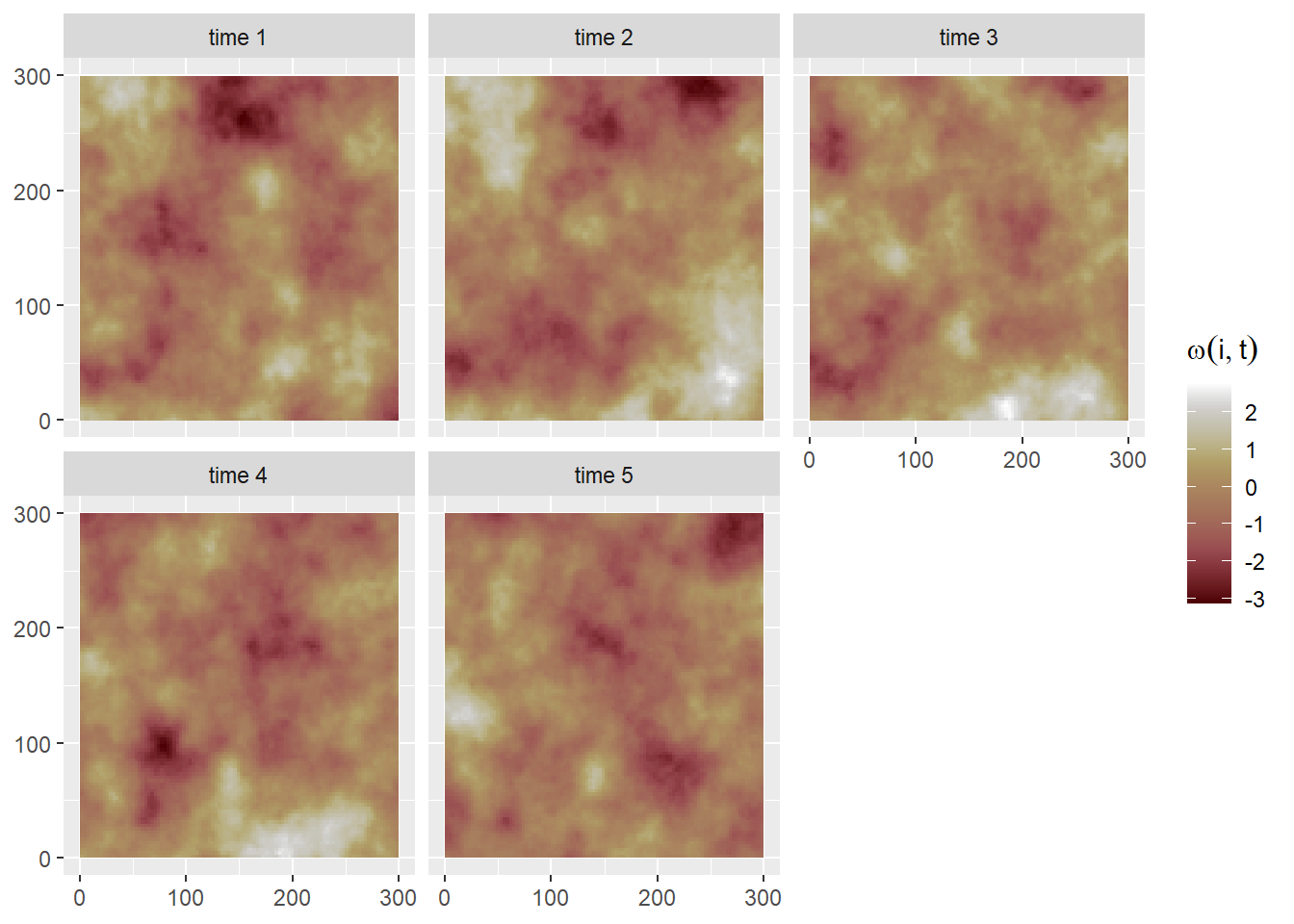
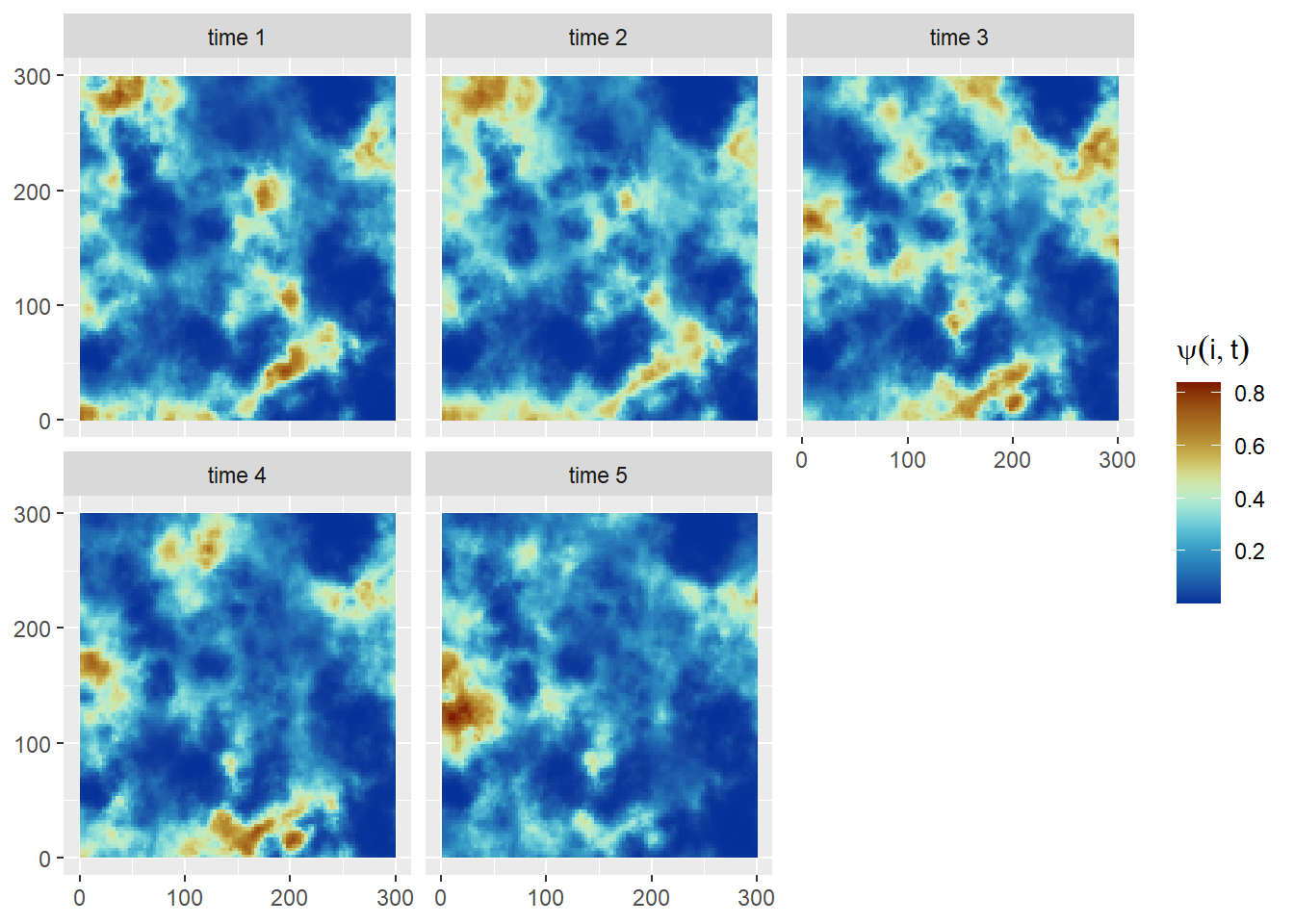
Then we can simulate the occupancy state of cell \(i\) at time \(t\) (Figure 6) as follows:
\[\begin{align}z_{it}&\sim \mathrm{Bernoulli}(\psi_{it})\nonumber\\\mathrm{logit}(\psi_{it}) &= \beta_0 + \beta_1 x^2_i + \omega_{it}.\end{align}\]Code
# State process model coeficcients
beta <- c(NA,NA)
beta[1] <- -0.5
beta[2] <- - 1
z.mat = psi.mat = matrix(NA,ncol=nT,nrow=dim(coord_grid)[1])
for(t in 1:nT){
# Occupancy probabilities
psi.mat[,t] = inla.link.logit(beta[1] + beta[2]*x_s^2 + omega_st[,t], inverse = T)
# True occupancy state
z.mat[,t] <- rbinom(ncells, size = 1, prob = psi.mat[,t])
}
Species detection/non-detection data at site \(i\) during visits \(j\) and time \(t\) is simulated from:
\[\begin{align} y_{ijt} &\sim \mathrm{Bernoulli}(p_{ijt} \times z_{it})\\ p_{ijt} &= \alpha_0 + \alpha_1 g_{ijt}\\ \end{align}\]Detection probabilities vary over space, time and survey through a simulated covariate \(g_{ijt}\). Often, locations are not surveyed regularly, resulting in unbalanced sampling scenarios. To simulate this, we introduce missing values with a constant probability for each site \(\times\) time \(\times\) survey combination. This missing values represent locations that were not surveyed on a given occasion.
Code
# Max number of vistis
K= 3
# Observational process model coeficcients
alpha <- c(NA,NA)
alpha[1] <- qlogis(0.3) # Base line detection probability
alpha[2] <- -1.5 # detection covariate effect
# Survey-level detection covariate g.t
g.t <- array(runif(n = nsites * K * nT, -1, 1), dim = c(nsites, K, nT))
# Observed data
# matrices to store each site x survey combination
y_mat <- p_mat <- matrix(NA,nrow = nsites,ncol=K)
colnames(y_mat) < paste("y",1:K,sep="")logical(0)Code
# empty list for storing each detection at each time point
y.t <- list()
# Constant NA probability
missing.p <- 0.1
# Detection/nondetection data (i.e. observed presence/absence records)
for(t in 1:nT){
for(j in 1:K){
p_mat[,j] <- inla.link.logit(alpha[1] +
alpha[2]*g.t[,j,t],
inverse = T)
y_mat[,j] <- rbinom(n = nsites,
size = 1,
prob = z.mat[site_id,t] * p_mat[,j])
turnNA <- rbinom(nsites, 1, missing.p)
y_mat[,j] <- ifelse(turnNA == 1, NA, y_mat[,j])
}
y.t[[t]] <- y_mat
}Finally, create and save data set.
Code
Obs_data.t <- do.call("rbind",y.t) %>% data.frame() %>%
bind_cols(apply(g.t,2L,c))
colnames(Obs_data.t) <- c(paste("y",1:K,sep=""),paste("g",1:K,sep=""))
Obs_data.t <- Obs_data.t %>%
mutate(cellid= rep(site_id,nT),time=rep(1:nT,each=nsites))
# centroid of the cell
Occ_data_2 <- suppressWarnings(customGrid %>%
st_centroid()) %>%
filter(sample==1) %>%
dplyr::select(-c('sample'))
Occ_data_2 <- left_join(Occ_data_2,Obs_data.t,by = "cellid")
# append coordinates as columns
Occ_data_2[,c('x.loc','y.loc')] <- st_coordinates(Occ_data_2)
Occ_data_2 = st_drop_geometry(Occ_data_2)
# Save CSV file for analysis
write.csv(Occ_data_2,file='Occ_data_2.csv',row.names = F)4 Spatially varying coefficients (SVC)
Here we simulate from a spatially varying coefficient model by adopting a space model with Matérn spatial covariance and a AR(1) time component. The same setting described for the space-time model will be used to drawn independent realization from the random field by using the book.rMatern()function. Then, temporal autocorrelation will be introduced to the dynamic regression coefficient \(\beta_t\) through the AR(1) structure described in the previous section (Note: The term \((1 - \rho^2)\) is added because of INLA internal AR(1) model parametrization; (Krainski et al. 2018)).
Code
# realization of the random field
set.seed(seed)
beta_1 <- book.rMatern(nT, coord_grid, range = range_spde,
sigma = sigma_spde)
# introduce temporal correlation
beta_1[, 1] <- beta_1[, 1] / (1 - rho^2)
for (t in 2:nT) {
beta_1[, t] <- beta_1[, t - 1] * rho + beta_1[, t] *
(1 - rho^2)
}Occupancy probabilities (Figure 7) are then defined on the logit scale as:
\[ \mathrm{logit}(\psi_{it}) = \beta_0 + \beta_{1}(i)t + \omega(i) \]
where \(\omega(i)\) is the same field defined for the simple spatial occupancy model (Section 2). The detection probabilities and observed occurrences are simulated in the same way as in Section 3.
Code
set.seed(seed)
# intercept and mean of the effect
beta0 <- -1
z.mat = psi.mat = matrix(NA,ncol=nT,nrow=dim(coord_grid)[1])
scale_t <- scale(1:nT) %>% c()
for(t in 1:nT){
# Occupancy probabilities
psi.mat[,t] = inla.link.logit(beta0 + beta_1[,t]*scale_t[t] + omega_s, inverse = T)
# True occupancy state
z.mat[,t] <- rbinom(ncells, size = 1, prob = psi.mat[,t])
}
# Max number of vistis
K= 3
# Observational process model coeficcients
alpha <- c(NA,NA)
alpha[1] <- qlogis(0.3) # Base line detection probability
alpha[2] <- -1.5 # detection covariate effect
# Survey-level detection covariate g2
g.t <- array(runif(n = nsites * K * nT, -1, 1), dim = c(nsites, K, nT))
# matrices to store each site x survey combination
y_mat <- p_mat <- matrix(NA,nrow = nsites,ncol=K)
# empty list for storing each detection at each time point
y.t <- list()
# Constant NA probability
missing.p <- 0.1
# Detection/nondetection data (i.e. observed presence/absence records)
for(t in 1:nT){
for(j in 1:K){
p_mat[,j] <- inla.link.logit(alpha[1] +
alpha[2]*g.t[,j,t],
inverse = T)
y_mat[,j] <- rbinom(n = nsites,
size = 1,
prob = z.mat[site_id,t] * p_mat[,j])
turnNA <- rbinom(nsites, 1, missing.p)
y_mat[,j] <- ifelse(turnNA == 1, NA, y_mat[,j])
}
y.t[[t]] <- y_mat
}
Obs_data.t <- do.call("rbind",y.t) %>% data.frame() %>%
bind_cols(apply(g.t,2L,c))
colnames(Obs_data.t) <- c(paste("y",1:K,sep=""),paste("g",1:K,sep=""))
Obs_data.t <- Obs_data.t %>%
mutate(cellid= rep(site_id,nT),time=rep(1:nT,each=nsites))
# centroid of the cell
Occ_data_3 <- suppressWarnings(customGrid %>%
st_centroid()) %>%
filter(sample==1) %>%
dplyr::select(-c('sample'))
Occ_data_3 <- left_join(Occ_data_3,Obs_data.t,by = "cellid")
# append coordinates as columns
Occ_data_3[,c('x.loc','y.loc')] <- st_coordinates(Occ_data_3)
Occ_data_3 = st_drop_geometry(Occ_data_3)
# Save CSV file for analysis
write.csv(Occ_data_3,file='Occ_data_3.csv',row.names = F)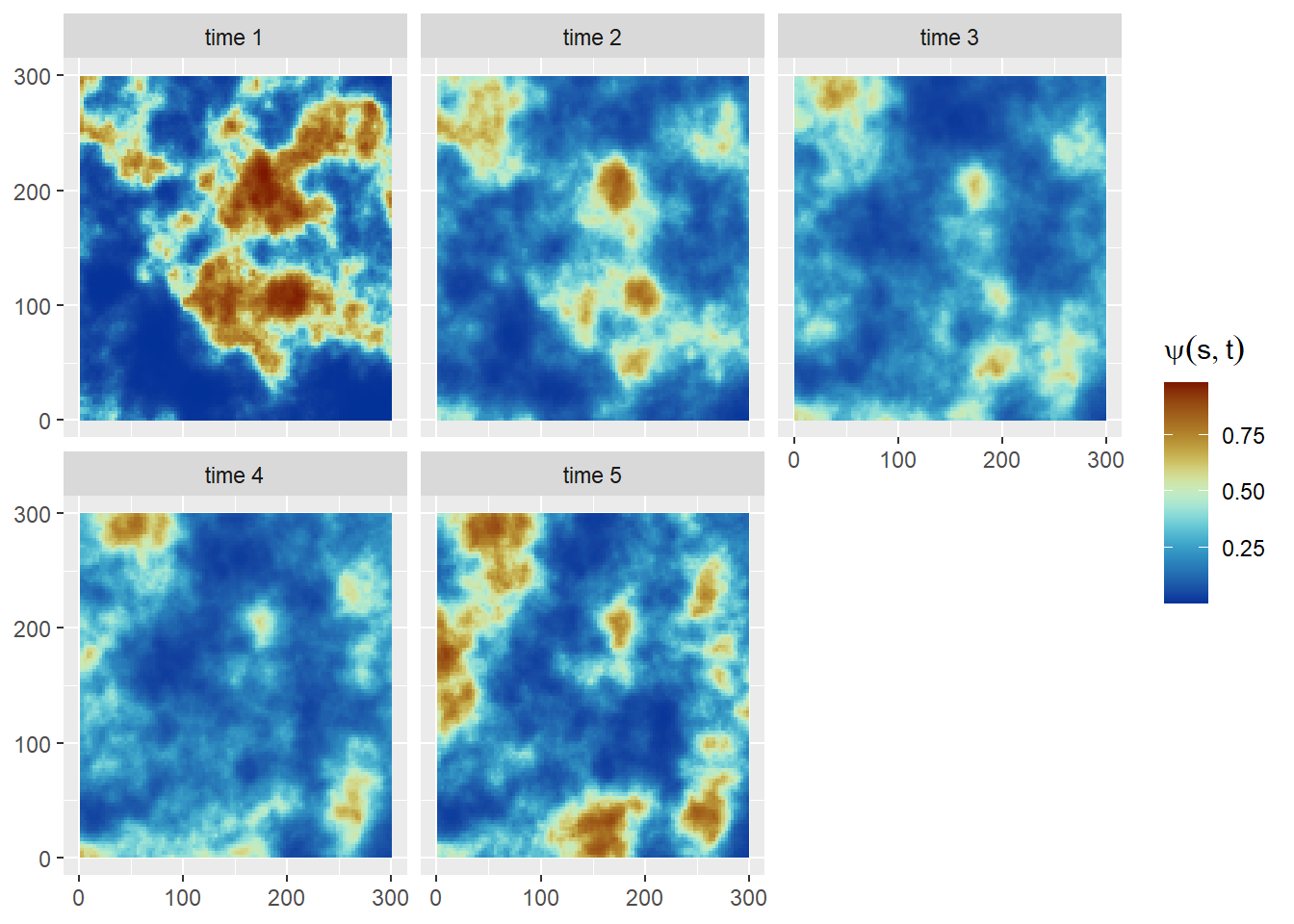
References
Footnotes
Note: If you are using INLA development version you might need modify the
book.rspde function. Specifically, change theinla.mesh.projectfunction tofmesher::fm_evaluatorfunction to project the mesh into the domain coordinates.↩︎